Model-Informed Drug Development Addresses COVID-19 Challenges

Drug developers know that the odds of anyone compound demonstrating safety and efficacy for a disease and its affected populations are low. How can drug developers improve these odds and increase the efficiency and effectiveness of drug development? One useful tool is model-informed drug development (MIDD), which uses computer models to inform the design of clinical trials or to run simulations when human or animal trials are not feasible. By ensuring that appropriate drugs are advanced and the clinical trial design is optimized, MIDD helps drug companies develop therapies for emerging diseases like COVID-19.
MIDD, also called modeling and simulation (M&S), is the application of in silico quantitative models in drug development to facilitate decision-making. MIDD is centered on knowledge and inferences generated from integrated mathematical models of the physicochemical characteristics of a molecule, its disposition in the body, and its mechanism of action, and how the drug might affect a disease from both an efficacy and a safety perspective. MIDD informs, reduces, and sometimes can eliminate the need for clinical trials, guiding decision-making on dose regimen optimization, safety, and toxicology, as well as clinical trial design and supportive evidence for efficacy.
Regulatory Framework
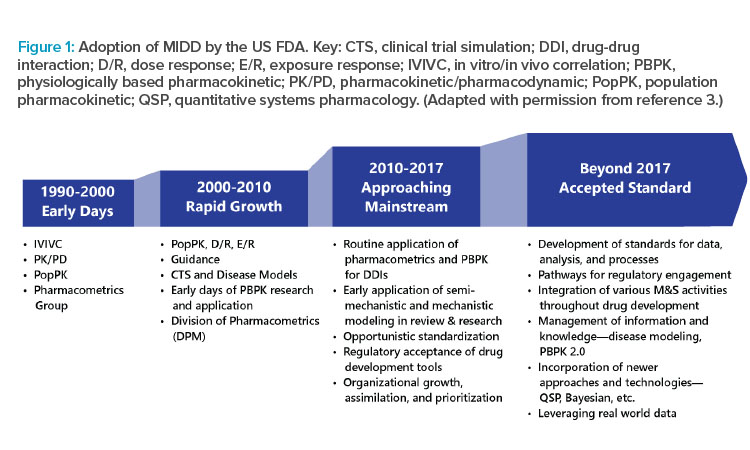
Encouraged by global regulators, MIDD is used in virtually all novel drug development programs. Specifically, the US FDA has MIDD provisions under the Prescription Drug User Fee Act (PDUFA) VI.1 One provision includes the MIDD Paired Meeting Pilot Program, where drug developers can meet with the FDA and receive input on their proposed MIDD application in specific programs.2 MIDD has become integrated as part of new drug applications (NDAs) and biologics license applications (BLAs) at the FDA (Figure 1).3 It is also accepted in applications to the European Medicines Agency and the Pharmaceuticals and Medical Devices Agency in Japan.1, 4
Work is ongoing to create international guidelines to standardize the validation and verification of MIDD models for use in regulatory filings. Two concerns in guideline development are (a) the reliability of models is only as good as the data used to generate them,1, 4 and (b) authorities do not want to limit sponsor creativity and innovation by making guidelines too restrictive.
Applications
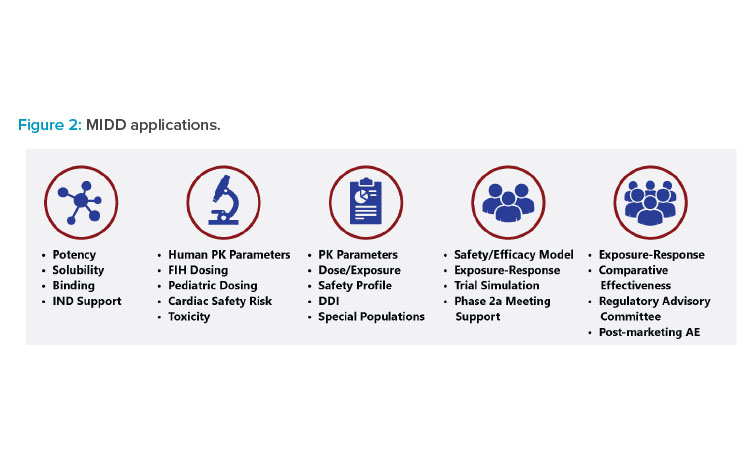
MIDD has many applications (Figure 2). It is used to determine product development go/no-go decisions, moving the most promising candidates forward and stopping development of prospects that are unlikely to be effective. MIDD is also employed to translate data from in vitro to in vivo research, animals to humans, smaller to larger/more diverse populations, adults to pediatric patients, and so on. Additionally, it can be used to model drug performance in different organs, evaluate drug-drug interactions (DDIs), and analyze optimal drug combinations.
Overall, MIDD helps drug developers answer key questions (Table 1). By leveraging expertise in clinical pharmacology, accelerated regulatory path-ways, and M&S, companies focusing on drug development can partner those with specific expertise in MIDD protocol development, thereby expediting trial enrollment.
MIDD is being used to aid in triaging COVID-19 drug candidates according to their likelihood of demonstrating safety and efficacy.
PBPK and POPPK Modeling
MIDD encompasses a range of techniques to inform drug development. Physiologically based pharmacokinetic (PBPK) and population pharmacokinetic (PopPK) modeling are two broadly used MIDD approaches. The former is a mechanistic, “bottom up” technique, and the latter is an empirical, “top down” technique.
PBPK M&S links in vitro data to in vivo absorption, distribution, metabolism, and excretion, as well as pharmacokinetic/pharmacodynamic (PK/PD) outcomes, to explore potential clinical complexities prior to human studies and support decision-making in drug development. Each individual subject has a set of PK/PD parameters based on their individual characteristics, drug concentration, and drug effect information. Thus, in individual model fitting, a full PK/PD profile is required to generate the PK/PD parameters of interest.
Population PK/PD M&S expands upon individual PK/PD analysis by (a) relating individual PK/PD parameters to a set of theoretical “typical” PK/PD parameters, and (b) quantifying the impact of known information (e.g., age, sex, weight, phenotype) on the variability in the individual PK/PD parameters.
| Topic | Questions |
|---|---|
| Selection of optimal drug candidates |
|
| Trial optimization |
|
| Safety and toxicity |
|
| Dose selection, special populations, and formulation |
|
| Regulatory and commercial success |
|
Development of COVID-19 Therapeutics
Several lines of investigation are open for COVID-19 drug development, and multifaceted modeling approaches are required to handle the different scenarios. MIDD is being used to aid in triaging COVID-19 drug candidates according to their likelihood of demonstrating safety and efficacy. Simulations are being performed to fill critical data gaps and determine in silico the impact of both new and existing therapies.
Determining Therapeutic Dosages
It is critical to optimize the dose for any therapy intended to treat COVID-19. If the dose is too low, it may be falsely concluded that the drug is ineffective. On the other hand, if a dose is too high, the drug may cause toxicity. Recent M&S work has predicted that certain drugs under consideration for COVID-19 treatment, ivermectin and lopinavir/ritonavir (LPV/r), are not efficacious at the doses studied.
Ivermectin
Ivermectin, an antiparasitic drug, is also approved for head lice and scabies. Researchers in Australia recently announced that a single dose of ivermectin could kill SARS-CoV2 in vitro within 24 hours.5 PBPK modeling was used to assess whether clinically relevant doses of ivermectin were able to attain the concentrations evaluated in vitro for treatment of patients with COVID-19.6 The simulations showed that ivermectin is unlikely to be effective. Even at a high-dose ivermectin regimen of 600 µg/kg daily for three days used in a previous clinical trial,7 with the most generous assumptions for clinical translation, the in vitro half-maximal inhibitory concentration (IC50) is more than 9-fold higher than the day 3 plasma-simulated maximum plasma concentration (Cmax), and more than 21-fold higher than the day 3 lung tissue–simulated Cmax. This dose scenario exceeds the highest regulatory-approved dose of ivermectin, a 200 µg/kg single dose for the treatment of strongyloidiasis (roundworm infection).8
If the developers of ivermectin create a potentially effective way of administering a higher dose of the drug, such as a nasal spray or inhaled drug, PBPK modeling could be applied to determine whether ivermectin in this form could effectively reduce viral counts. However, if the higher dose of ivermectin is limited because of safety concerns, it is unlikely that this candidate will move forward.
LPV/r
The limited viral kinetic data available for COVID-19 suggest that the duration of viral shedding (i.e., the process of the virus being released into the environment after it has replicated inside the body) is longer than that for influenza, and longer treatment may be necessary to continue to fight the virus. Investigators are interested in LPV/r, an approved HIV treatment, as a potential COVID-19 therapy because it decreases HIV levels in blood and theoretically may have similar effects on SARS-COV-2.9
In vitro data on the effects of LPV/r against coronaviruses indicate that the drug combination may be significantly less active against SARS-COV-2 compared to HIV-1.9 This suggests that the HIV dose for LPV/r would be unlikely to be successful against SARS-COV-2, where it is much less potent.
A collaborative research team used PopPK models and tested a range of potential PK/PD relationships to simulate LPV/r plasma and lung exposures in adults.10 The analysis indicates that standard LPV/r doses are at high risk of COVID-19 treatment failure, especially without a loading dose. The standard LPV/r dose takes 36–48 hours to reach full plasma concentration, which would lead to a loss of valuable time in the SARS-COV-2 treatment window. Given that the dose regimen for COVID-19 needs to reach therapeutic concentrations quickly, the models indicate that a standard HIV dosing regimen is unlikely to be effective against SARS-COV-2.10
Additional modeling with a loading dose could be performed to determine whether reaching higher plasma LPV/r concentrations in a shorter time can alter the COVID-19 disease course (e.g., whether the patient requires hospitalization and a ventilator).
Viral Kinetic Modeling
Another approach for advancing treatments for COVID-19 involves viral kinetic models that use mathematical equations to describe how viral load changes with time in an infected patient.10 Originally used for HIV-1, these models can provide important information about the cell infection rate, viral production, and clearance rates. They are also used to predict disease course and treatment outcomes. With SARS-COV-2, there is a wide range of potential disease courses, from asymptomatic cases to patients requiring hospitalization and a ventilator.11 Therefore, COVID-19 models may include additional variables to account for disease course variability and complexity.
Research teams are combining viral kinetic modeling with PK/PD modeling to quantify drug effects based on their mechanism of action and evaluate in silico the efficacy of specific drug combinations in treating COVID-19 and other viral diseases.12 These models can help link the antiviral drug of interest to its impact on a patient’s viral load, which is an important driver of the duration and severity of disease. Patients who shed virus are infectious, and such viral kinetic models have been coupled to epidemiological models to assess the effectiveness of drug treatment strategies on reducing the number of infections in a population (i.e., “flattening the curve”).
Lung Exposure Modeling
Another MIDD approach relevant to COVID-19 involves modeling the lung exposure of drugs. To combat the tuberculosis (TB) epidemic, a research team in collaboration with several leading global health institutions developed a multicompartment permeability-limited lung model. This newly developed PBPK model helps drug developers leverage in vitro and in silico data to understand drug disposition and penetration in plasma, lung tissue, epithelial lining fluid, and TB granulomas. In addition, these tools allow researchers to simulate a range of variables—drug dose, disease state, and concomitant medications—and thus support designing more-effective drug regimens.13 This model has been used to evaluate drugs that can treat COVID-19.14
Conclusion
The global community of scientists is working 24/7 to identify, study, and test potential treatments and cures for many diseases, including COVID-19. In the current pandemic, there is a compelling need to speed progress, evaluate opportunities to shift regulatory paradigms, and efficiently make go/no-go decisions. It is time for researchers to open the MIDD toolbox and leverage it. Together, using multiple advanced modeling tools, collaboration, and expertise, the scientific community will prevail in finding appropriate treatments and preventive measures for COVID-19.
Acknowledgment
The authors thank Mia DeFino, MS, ELS, for medical writing support.



